Forensic Anthropology Vol. 2, No. 2: 121–128
DOI: 10.5744/fa.2019.1019
TECHNICAL NOTE
Using Biological Profile Data to Inform a DNA Sequencing Strategy
Carrie Browna* ● Jeffrey James Lyncha
ABSTRACT: Frequency distributions of antemortem stature and age for individuals who were casualties onboard the USS Oklahoma are visually compared to frequency distributions of point estimates for long bone stature and pubic symphysis age drawn from the large commingled assemblage that is associated with this loss incident. Based on similarities in the means and standard deviations of these distributions, a four-tiered DNA sequencing strategy is devised to prioritize the sampling of elements that are at least one standard deviation from Oklahoma antemortem mean age and/or stature. The rationale for this approach is that elements providing estimates that are at least one standard deviation from antemortem stature and/or age means are more likely to be from individuals who also fall at least one standard deviation from the means for one or both of these categories. This prioritization strategy resulted in nearly doubling identifications during the initial phases of the project. This success demonstrates the ability to use biological profile data to aid in the DNA sequencing process and the importance of continued interdisciplinary work in resolving commingled assemblages.
KEYWORDS: forensic anthropology, resolution of commingling, mitochondrial DNA analyses, stature, age
Large commingled assemblages present unique challenges for individual identification, because the segregation of remains into discrete individuals can be difficult and time-consuming. These assemblages can be problematic from data management and analytical perspectives, and often an increase in the number of casualties leads to a decrease in the discriminatory power of anthropological techniques such as visual pair matching and articulation (SWGANTH 2013). These challenges can lead to lengthy analytical timeframes and delays in identification, and a multidisciplinary approach to resolving commingling is both espoused and practiced (e.g., Salado Puerto et al. 2014; SWGANTH 2013).
Skeletal and DNA analyses are commonly employed together in cases of commingling (e.g., Damann & Edson 2008; Finlayson et al. 2017; Hines et al. 2014; Mundorff et al. 2014; Salado Puerto et al. 2014). One of the first steps in resolving commingling is for the anthropologist to do as much sorting as possible—via pair matching, articulation, and the biological profile—in order to determine how many individuals are present and then segregate those individuals (Damann & Edson 2008). Provenience and taphonomic evidence should also be used to aid in this process, as applicable and available. Even in approaches that rely heavily on DNA, the anthropologist maintains an important role in making and verifying associations of elements, detecting and resolving commingling pre- and post-sampling, confirming biological profile data, and reducing the cost and burden of DNA analysis by associating elements (Damann & Edson 2008; Yazedijian & Kešetović 2008).
Part of the skeletal sorting strategy entails relying on inter-individual differences in size, morphology, and age to segregate skeletal elements. It is thus important to consider the demographic composition of the loss population and the assemblage, as this can be a significant factor in the resolution of commingling. Generally, when faced with a homogeneous group (i.e., composed of individuals of the same sex and similar age, ancestry, and stature) it is more difficult to sort using skeletal techniques. However, as Yazedijian and Kešetović (2008:280) find, even in a sample composed primarily of “men of fighting age” from a single ancestral background where stature is normally distributed, it is still possible to separate outliers.
In cases where remains are extensively commingled, fully skeletonized, disarticulated, and/or lacking provenience or other recovery information, or where the anthropologist is otherwise hindered in sorting remains prior to DNA sampling, DNA analysis may be relied on heavily to aid in the segregation of skeletal elements. In these instances, DNA analysis has the potential to be time-consuming and costly. What then becomes the best strategy to approach the analysis of numerous samples when individuals cannot be reliably segregated prior to DNA sequencing? Simply sequencing the samples by order of receipt may not represent the best strategy.
Other researchers have provided information on the preservation of DNA by element (e.g., Damann & Edson 2008; Edson et al. 2004; Hansen et al. 2017; Hines et al. 2014; Mundorff et al. 2009; Pinhasi et al. 2015), which is useful in terms of what elements should be given priority for DNA sampling but not for the order in which they should be sequenced once sampled. Currently, there is no known research that addresses how to prioritize samples for sequencing in large, heavily commingled assemblages that cannot be reliably segregated prior to DNA analysis. Having a strategy to prioritize DNA analyses is potentially useful to maximize identifications in the early stages of an ongoing project where DNA is crucial to those identifications. Rather than waiting for all DNA to be sequenced, samples can be prioritized based on which are more likely to be associated with certain individuals (e.g., very tall or very short individuals). In long-term projects where a significant amount of time has passed since the deaths of the individuals, there are fewer surviving family members with each passing year and identifications become a race against time. Thus, making identifications quickly is often important. A prioritization strategy may also be beneficial for decreasing the overall number of samples requiring sequencing. As outliers are identified first, sampled elements may be able to be pair matched and DNA processing halted for those elements, but it is important to do this in tandem with the DNA laboratory.
This study compares antemortem and postmortem data from a commingled assemblage with a historically known size (i.e., all casualties from the Oklahoma). Based on this investigation and on age and stature variation in the population, it then outlines an effective and demographically informed strategy to prioritize DNA sequencing of postcranial elements using antemortem age and stature data. This strategy enables the identification of individuals in this assemblage prior to completion of all DNA analyses, reducing the pool of missing persons and the time families wait for notification.
Materials and Methods
This study employs skeletal elements and mitochondrial DNA (mtDNA) results from the USS Oklahoma assemblage. For information on the assemblage and analyses, including the extent of commingling, see Brown (2019). The identification count given here includes only those individuals with postcranial elements that were identified or in analysis as of 1 June 2018 (n = 93), and it does not include any identifications prior to project initiation in 2015.
During the inventory process, standard and supplemental measurements were taken for all elements, as applicable (Byrd & Adams 2003; Byrd & LeGarde 2014; Moore-Jansen et al. 19941), and long bone epiphyseal fusion and pubic symphyses were scored following McKern and Stewart (1957). Only those elements that were sampled for DNA analyses were considered in this study, and those elements that did not have maximum length measurements or the ability to assess age via fusion or the pubic symphysis were excluded. All cranial and dental samples were sequenced prior to the postcranial elements to aid with MNI estimation and dental identifications, and thus they also are excluded from this study.
For the long bones—humerus, radius, ulna, femur, tibia, and fibula—stature point estimates were calculated by element using linear regression equations from Trotter’s Black and White Male combined data set in FORDISC 3 (Jantz & Ousley 2005). This process was automated in OsteoSort (2016) in order to create frequency distributions of point estimates by element, pooling the left and right sides. A frequency distribution was also created for the mean ages (point estimates) per pubic symphysis composite score, and again the sides were pooled. No distributions were created for long bone epiphyseal fusion, as the method following McKern and Stewart (1957) only provides age intervals by epiphysis per stage of fusion (e.g., proximal humerus, Stage 0, observed from 17 to 20 years). Descriptive statistics for postmortem estimates providing mean ages were calculated in R (R Core Team 2014).
Antemortem ages and statures were drawn from military records. When more than one stature was listed in the records, an average of reported statures was produced and used for comparative purposes. Frequency distributions and descriptive statistics for antemortem stature and age were calculated in R (R Core Team 2014).
Antemortem and postmortem distributions were compared visually and using descriptive statistics (e.g., similar means and standard deviations). This formed the basis for the tiered prioritization strategy of the postcranial samples submitted for mtDNA analyses (See Results, below). All samples were sent to the Armed Forces DNA Identification Laboratory (AFDIL) for sequencing.
Results
Antemortem stature and age distributions are given in Figures 1 and 2; postmortem stature point estimate distributions for all six limb bones and pubic symphysis age distributions are given in Figures 3 through 9. Table 1 provides the mean, standard deviation, and within and outside one standard deviation intervals per distribution.
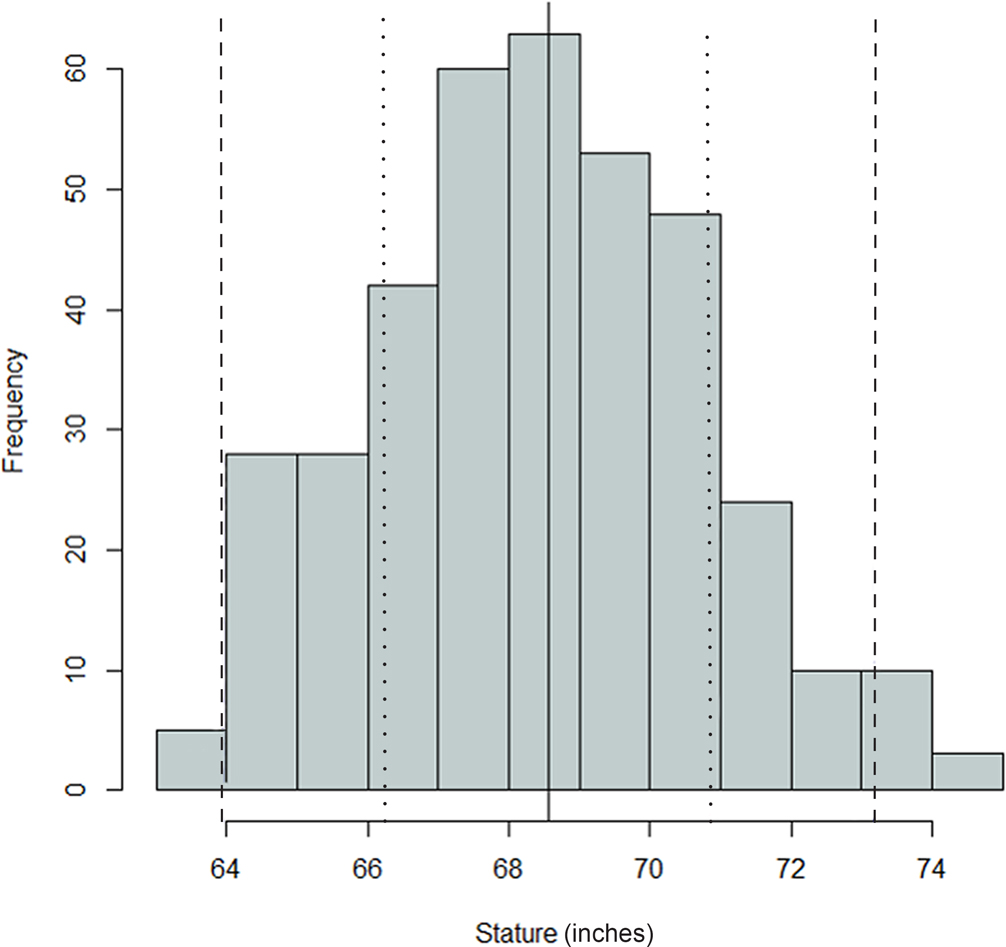
FIG. 1—Distribution of antemortem statures for the Oklahoma sample; mean = 68.56 inches, SD = 2.31 inches, n = 374. The black, dashed, and dotted lines are mean, one standard deviation, and two standard deviations, respectively.
FIG. 2—Distribution of antemortem ages for the Oklahoma sample; mean = 24.49 years, SD = 6.4 years, n = 394. The dashed line indicates the median, and the solid line indicates the mean.
FIG. 3—Distribution of stature point estimates for the humerus; mean = 68.26 inches, SD = 1.86 inches, n = 507. The solid, dashed, and dotted lines are mean, one standard deviation, and two standard deviations, respectively.
FIG. 4—Distribution of stature point estimates for the ulna; mean = 68.35 inches, standard deviation = 1.49 inches, n = 328. The solid, dashed, and dotted lines are mean, one standard deviation, and two standard deviations, respectively.
Comparing Figures 1 through 9, there is a clear similarity between distributions of antemortem and postmortem stature and age, though it is noted that the use of McKern and Stewart (1957) demonstrates a regression to the mean for pubic symphysis age point estimates as compared to the antemortem data. Additionally, the average of tibiae point estimates is an inch shorter than the averages for all other long bones in the postmortem assemblage and for the average antemortem stature of Oklahoma crew members. However, because the standard deviation for stature based on the tibia point estimates is similar to the other long bones and the distribution of point estimates based on the tibia is normal, the tibia data are still used in the prioritization strategy. Because of the similar standard deviations, it was determined that the issue is with how condylo-malleolar length is being translated into stature and not because of great variation in tibia length.2 Of 394 individuals, 160 are at least one standard deviation from the antemortem mean stature and/or age; this represents 40.6% of the Oklahoma loss population that can potentially be segregated based on stature and/or age.
FIG. 5—Distribution of stature point estimates for the radius; mean = 68.44 inches, SD = 1.47 inches, n = 325. The solid, dashed, and dotted lines are mean, one standard deviation, and two standard deviations, respectively.
FIG. 6—Distribution of stature point estimates for the femur; mean = 68.17 inches, SD = 1.97 inches, n = 642. The solid, dashed, and dotted lines are mean, one standard deviation, and two standard deviations, respectively.
FIG. 7—Distribution of stature point estimates for the tibia; mean = 67.08 inches, SD = 1.88 inches, n = 573. The solid, dashed, and dotted lines are mean, one standard deviation, and two standard deviations, respectively.
FIG. 8—Distribution of stature point estimates for the fibula; mean = 68.45 inches, SD = 1.97 inches, n = 360. The solid, dashed, and dotted lines are mean, one standard deviation, and two standard deviations, respectively.
FIG. 9—Distribution of age estimates based on the public symphysis; mean = 24.31 years, SD = 5.21 years, n = 405. The dashed line indicates the median, and the solid line indicates the mean.
It is important to note that biological profile characteristics are not the only means by which segregation occurs in the Oklahoma project. Dental records, the presence of a DNA reference sample, and the commonality of the mtDNA sequence are also important factors in segregation and eventual identification. For example, an individual may be of average height and age but have a rare mtDNA sequence and thus is easily segregated and identified because no other individuals share that mtDNA sequence, while an individual who is an outlier for age and/or stature has no DNA reference on file or a common DNA sequence and thus takes longer to segregate and identify. For additional information on DNA reference samples and the identification process for Oklahoma crew members, see Brown (2019). The prioritization strategy does not address the overall identification success for the project, which is unknown at this time, but outlines a method by which elements can be moved forward in the DNA sequencing queue using height and age estimations, rather than waiting for all DNA samples to be sequenced by order of receipt. It is important to note that due to the extensive commingling in this assemblage, very few of the identifications are made without DNA data.
Based on the composition of the loss population and the commingled assemblage and similarities in means and standard deviations, postcranial samples were prioritized in four tiers, representing 32.6% of the total samples submitted to AFDIL (Table 2). The prioritization strategy targeted elements with stature and/or age estimates that fell at least one standard deviation from the Oklahoma (antemortem) means. The rationale was to prioritize elements that were more likely to come from individuals at least one standard deviation from antemortem mean height and/or age, as these elements are the easiest to segregate from the commingled assemblage; however, the strategy was not find elements that belong to the same individual. The first tier to be prioritized was long bones with stature point estimates greater than or less than two standard deviations from the point estimate mean per element, as these likely represented the tallest and shortest individuals in the assemblage, respectively. The second tier included those elements that produced age estimates less than or equal to 20 years and greater than or equal to 30 years (Stage 0, 1, and 2 fusion for the long bones, and composite scores of 0–5 and 14–15 for the pubic symphysis), in order to capture those individuals at or outside one standard deviation from the antemortem mean age. The third tier consisted of elements exhibiting Stage 3 fusion, as this generally occurs in the early twenties, and the fourth tier was long bones providing point estimates between one and two standard deviations from the point estimate mean per element. This particular order was chosen to attempt to sequence elements from individuals furthest from the mean first and then progress through the remains from greatest to least deviation from the mean. Due to the antemortem age distribution for the loss population, epiphyses fusing in the mid- to late twenties were not prioritized as part of the strategy. If an element was placed in multiple levels (e.g., femur with unfused epiphyses and point estimate of 72 inches), it was given the higher of the ranked priorities.
TABLE 1—Descriptive statistics for antemortem and postmortem stature and age distributions, including within and outside one standard deviation intervals.
|
Biological parameter |
Mean |
Standard Deviation |
Within 1 Standard Deviation |
Outside 1 Standard Deviation |
||||
|
Antemortem stature (inches) |
68.56 |
2.31 |
66.25–70.87 |
<66.25; >70.87 |
||||
|
Postmortem stature (inches)—humerus |
68.26 |
1.86 |
66.40–70.12 |
<66.40; >70.12 |
||||
|
Postmortem stature (inches)—ulna |
68.35 |
1.49 |
66.86–69.84 |
<66.86; >69.84 |
||||
|
Postmortem stature (inches)—radius |
68.44 |
1.47 |
66.97–69.91 |
<66.97; >69.91 |
||||
|
Postmortem stature (inches)—femur |
68.17 |
1.97 |
66.20–70.14 |
<66.20; >70.14 |
||||
|
Postmortem stature (inches)—tibia |
67.08 |
1.88 |
65.20–68.96 |
<65.20; >68.96 |
||||
|
Postmortem stature (inches)—fibula |
68.45 |
1.97 |
66.48–70.42 |
<66.48; >70.42 |
||||
|
Antemortem age (years) |
24.49 |
6.40 |
18.09–30.89 |
<18.09; >30.89 |
||||
|
Postmortem age (years)—pubic symphysis |
24.31 |
5.21 |
19.10–29.52 |
<19.10; >29.52 |
TABLE 2—Prioritization of samples: descriptions and counts. Total DNA samples submitted from June 2015 to October 2017, N = 4,749.
|
Level |
Description |
n |
% of total |
|||
|
0 |
All cranial and dental |
604 |
12.7 |
|||
|
1 |
Stature point estimate ± 2 standard deviations (in inches) |
89 |
1.9 |
|||
|
2 |
Age estimate ≤ 20, ≥ 30 (in years) |
247 |
5.2 |
|||
|
3 |
Age estimate ~21 (in years) |
106 |
2.2 |
|||
|
4 |
Stature point estimate ± 1–2 standard deviations (in inches) |
501 |
10.5 |
|||
|
Total |
All prioritized |
1,547 |
32.6 |
As of 1 June 2018, 44 individuals of the 93 individuals identified, approved for identification, or currently segregated from the assemblage and undergoing analysis were at least one standard deviation from mean stature and/or age. This represents 47.3% of the study sample. Figure 10 displays the breakdown of these 44 by status: identified, approved for identification, or currently undergoing analysis and segregated from the commingled assemblage. These 44 individuals represent 27.5% of the 160 Oklahoma casualties that are at least one standard deviation from mean stature and/or age.
The prioritization strategy resulted in the mtDNA sequencing of 943 postcranial samples. When combined with the cranial and dental samples that were processed first, this results in a total of 1,547 samples processed as priority, out of a total 4,749 submitted (32.6%). The completion of the postcranial priority tiers took approximately 18 months, though it should be noted that during this time AFDIL also processed many other cases and projects as well as additional nuclear testing for the Oklahoma project.
FIG. 10—Identification and analysis of individuals at least one standard deviation from antemortem stature and age means compared to individuals similar to the mean.
Discussion
As of 1 June 2018, mtDNA sequencing was just over 65% complete for the entire project, and all priority samples were completed. Project analyses have resulted in the identification or segregation of 93 individuals, and nearly half of them are at least one standard deviation from the loss population mean antemortem stature and/or age. This indicates that the biological-profile-informed strategy to target specific samples for sequencing, rather than relying on order of submission, can be considered a success in this assemblage in terms of segregating and identifying individuals earlier than if waiting for all DNA to be sequenced, because it resulted in nearly double the number of identifications in the initial phase of the project. This strategy does not consider overall project success in terms of identifications, since the project is ongoing, and identifications rely heavily on DNA. Due to this reliance on DNA, ultimately identification is related largely to having a reference sample on file and the relative rarity of that sequence in the Oklahoma sample as well as the ability to use other methods to segregate and associate elements and individuals.
Biological anthropologists use their knowledge of human variation to segregate individuals. In commingled assemblages this variation enables anthropologists to employ smart sampling strategies that can reduce the burden on the DNA testing laboratory by associating elements prior to DNA analyses. In this case, given the size of the assemblage and high degree of commingling, sorting was not possible prior to DNA testing, so variations in size and morphology were employed to inform the order of sequencing. Additionally, once an element on a priority list has been sequenced, analysts worked to associate contralateral elements via osteometric sorting (Byrd & Adams 2003; Byrd & LeGarde 2014) and visual pair matching in order to reduce the burden on AFDIL by canceling elements that were sampled but subsequently successfully associated to sequenced elements.
The prioritization strategy employed here has not resulted in a dramatic decrease in the overall number of samples requiring analysis at AFDIL. However, even several dozen cancellations enables AFDIL to put time and resources toward other samples. Where this process likely will provide some efficiency is in the association of additional remains to previously identified individuals. Because identifications are made prior to all DNA samples being sequenced, there are instances in which an element or elements are associated to an individual following identification. In these cases, additional skeletal, DNA, and dental reports must be written, as applicable, in order to associate the additional remains. By prioritizing the sequencing of elements that are likely from individuals who are outside one standard deviation in height and/or age, it is more likely that all elements for a single individual will be sequenced first, then segregated, and, finally, identified at the same time, thus reducing the workload involved in writing multiple reports later. However, this efficiency has not yet been proven or disproven in the Oklahoma project.
Prioritizing the samples is important given the five-year timeline for the Oklahoma project and the amount of time that has passed since the incident. While prioritizing likely will not affect the overall number of identifications, it is important in terms of identifying individuals earlier on, thereby reducing the missing persons pool. Identification now, rather than once all DNA has been processed, is also important for family members. December 2018 marked the 77th anniversary of the bombing of Pearl Harbor and while there are still living siblings of the Oklahoma crew members, there are fewer with each passing year.
By employing this strategy, an inherent statement is being made about the usefulness of each element in terms of segregation potential. In this study, elements that do not appear in the priority list are those that do not have information that is considered probative to distinguishing individuals based on the distribution of age and/or stature in the loss population. However, as detailed above, this does not necessarily affect the ultimate identification, as other lines of evidence are also used to support the association of a set of segregated remains to an individual. But, the prioritization strategy can and should be customized based on the demographic composition of the loss population. For example, ancestry was not used for this project, because all crania were sequenced prior to the development of the tiered list. However, had this not been the case, ancestry assessment of the crania would have been included in the tiers, since the Oklahoma loss population included only a small number of individuals of Asian and African ancestry that would likely have been able to be segregated based on morphology and osteometry.
The prioritization strategy outlined here relies on standardized data collection of the entire assemblage. This step is crucial because it provides the baseline for understanding the assemblage in terms of demographic composition and number of individuals represented. In projects where multiple analysts are collecting data on the skeletal remains, it is also important to outline standard protocols, such as what age estimation methods are used, so that data collection is consistent across the assemblage.
Additional limitations to this method include the availability of antemortem anthropological data, the preservation of skeletal elements, and the recovery of elements. The antemortem and postmortem frequency distributions are only as good as the data available. If biological profile data for the loss population are not available or measurements and estimates cannot be made on the remains due to preservation or absence of elements, it is not possible to create frequency distributions and sort individuals or elements using this strategy.
The use of the McKern and Stewart (1957) pubic symphysis ages demonstrates a well-known phenomenon in biological anthropology: age mimicry of the study sample to the method reference sample (Bocquet-Appel & Masset 1982). While the Oklahoma assemblage is theoretically similar to the McKern and Stewart (1957) sample—young, White military males—the samples are not identical. When the McKern and Stewart (1957) pubic symphysis is used, it underages the upper end of the Oklahoma distribution, pushing individuals closer to the sample mean.
Finally, this study demonstrates that interdisciplinary work continues to be vital to the resolution of commingling. DNA and skeletal analysis should be used in tandem, in a type of feedback loop where each informs the other. In this case, working relationships between anthropologists and DNA analysts are vital to the success of the prioritization strategy as well as the project as a whole.
Conclusion
This study demonstrates the utility of using demographic data to prioritize the DNA sequencing of elements in a large commingled assemblage. This research focused on isolating potential outliers in both the antemortem and postmortem data and focusing on these individuals for earlier segregations and identifications. Because of the demographic makeup of the loss population, the clavicle and innominates were not targeted in the initial priority sequencing strategy. However, they have now been added as the fifth tier in order to aid with age estimates during the association to known individuals phase of the project.
References
Bocquet-Appel J-P, Masset C. Farewell to paleodemography. Journal of Human Evolution 1982;11(4):321–333.
Brown CA. The USS Oklahoma identification project. Forensic Anthropology 2019;2(2):102–112.
Byrd JE, Adams BJ. Osteometric sorting of commingled human remains. Journal of Forensic Sciences 2003;48(4):717–724.
Byrd JE, LeGarde C. Osteometric sorting. In: Adams BJ, Byrd JE, eds. Commingled Human Remains: Methods in Recovery, Analysis, and Identification. San Diego: Academic Press; 2014:167–192.
Damann FE, Edson SM. Sorting and identifying commingled remains of U.S. war dead: The collaborative roles of JPAC and AFDIL. In: Adams BJ, Byrd JE, eds. Recovery, Analysis, and Identification of Commingled Human Remains. Totowa, NJ: Humana Press; 2008:301–316.
Edson SM, Ross JP, Coble MD, Parsons TJ, Barritt SM. Naming the dead—Confronting the realities of the rapid identification of degraded skeletal remains. Forensic Science Review 2004;16(1):63–90.
Finlayson JE, Bartelink EJ, Perrone A, Dalton K. Multimethod resolution of a small-scale case of commingling. Journal of Forensic Sciences 2017;62(2):493–497.
Hansen HB, Damgaard PB, Margaryan A, Stenderup J, Lynnerup N, Willerslev E, et al. Comparing ancient DNA preservation in petrous bone and tooth cementum. PLoS One 2017;12(1):e0170940.
Hines DZC, Vennemeyer M, Amory S, Huel RLM, Hanson I, Katzmarzyk C, et al. Prioritized sampling of bone and teeth for DNA analysis in commingled cases. In: Adams BJ, Byrd JE, eds. Commingled Human Remains: Methods in Recovery, Analysis, and Identification. San Diego: Academic Press; 2014:275–305.
Jantz RL, Ousley SD. FORDISC 3.0: Personal computer forensic discriminant functions. University of Tennessee, Knoxville; 2005.
Langley NR, Jantz LM, Ousley SD, Jantz RL, Milner G. Data collection procedures for forensic skeletal material 2.0. University of Tennessee, Knoxville; 2016
Lynch JJ, Brown C, Palmiotto A, Maijanen H, Damann F. Reanalysis of the Trotter tibia quandary and its continued effect on stature estimation of past-conflict service members. Journal of Forensic Sciences 2019;64(1):171–174.
McKern TW, Stewart, TD. Skeletal age changes in young American males: Analyzed from the standpoint of age identification. Report EP-45. Natick, MA: Quartermaster Research and Development Command and Development Center, Environmental Protection Research Division; 1957.
Moore-Jansen PM, Ousley SD, Jantz RL. Data collection procedures for forensic skeletal material. Report of Investigations No. 48. University of Tennessee, Knoxville; 1994.
Mundorff AZ, Bartelink EJ, Mar-Cash E. DNA preservation in skeletal elements from the World Trade Center disaster: Recommendations for mass fatality management. Journal of Forensic Sciences 2009;54(4):739–745.
Mundorff A, Shaler R, Bieschke ET, Mar-Cash E. Marrying anthropology and DNA: Essential for solving complex commingling problems in cases of extreme fragmentation. In: Adams BJ, Byrd JE, eds. Commingled Human Remains: Methods in Recovery, Analysis, and Identification. San Diego: Academic Press; 2014:257–273.
OsteoSort. https://osteocoder.com/. Accessed October 12, 2016.
Pinhasi R, Fernandes D, Sirak K, Novak M, Connell S, Alpaslan-Roodenberg S, et al. Optimal ancient DNA yields from the inner ear part of the human petrous bone. PLoS ONE 2015;10(6):e0129102.
R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria, 2014. http://www.R-project.org/. Accessed January 30, 2018.
Salado Puerto M, Egaña S, Doretti M, Vullo CM. A multidisciplinary approach to commingled remains analysis: Anthropology, genetics, and background information. In: Adams BJ, Byrd JE, eds. Commingled Human Remains: Methods in Recovery, Analysis, and Identification. San Diego: Academic Press; 2014:307–335.
SWGANTH. Resolving commingled human remains. Revision 2. Available at: https://www.nist.gov/topics/forensic-science/anthropology-subcommittee. Issued January 22, 2013. Accessed May 29, 2018.
Yazedijian L, Kešetović R. The application of traditional anthropological methods in a DNA-led identification project. In: Adams BJ, Byrd JE, eds. Recovery, Analysis, and Identification of Commingled Human Remains. Totowa, NJ: Humana Press; 2008:271–284.
1. At the time data collection started for the Oklahoma project, the updated data collection procedures (Langley et al. 2016) had not yet been released, so all measurements were taken following the Moore-Jansen et al. (1994) standards for consistency throughout the duration of the project.
2. For a detailed analysis of this issue, see Lynch et al. (2019). The condylo-malleolar length of the tibia was measured following the description in Moore-Jansen et al. (1994).
aDefense POW/MIA Accounting Agency—Laboratory, Offutt AFB, NE, USA
*Correspondence to: Carrie Brown, Defense POW/MIA Accounting Agency—Laboratory, 106 Peacekeeper Dr., Bldg. 301, Offutt AFB, NE 68113, USA
E-mail: carrie.a.brown40.civ@mail.mil
Received 11 June 2018; Revised 28 October 2018; Accepted 14 January 2019